Journal of Animal Research: v.10 n.1, p. 143-148. February 2020

DOI: 10.30954/2277-940X.01.2020.21
Isolation and Molecular Characterization of Extended Spectrum Beta Lactamase Producing Escherichia coli from Milk
ABSTRACT
Milk plays a major role as a source of nutrition in the diet but contaminated milk can be a source of harmful bacteria. Escherichia coli is opportunistic pathogen and is responsible for a wide range of infections. The prevalence of pathogenic multi-drug resistant extended-spectrum β-lactamase (ESBL)-producing E. coli is increasing and becoming a global concern. A study was carried out to isolate ESBL producing E. coli from 150 milk samples from Anand and around villages. Total 94(62.66%) samples were found positive as E. coli by isolation on MacConkey and Eosin Methylene Blue agar which were confirmed by primary & biochemical tests including Gram’s staining. Antibiotic sensitivity test (ABST) was performed against 6 antibiotics and isolates found resistant to Aztrionem: 58(61%), Cefoxitin: 20(21%), Ceftriaxone: 56(59%), Ceftazidime: 62(65%), Cefpodoxime: 34(44.73%) & Ceftazidime + Clavulanic acid: 8(8.5%). A total 34(36.17%) ESBL producing E. coliwere phenotypically confirmed by ABST and Epsilometer test. Genotypic confirmation of 34 isolates was done by PCR and isolates found positive for bla CTX M3 gene: 18(52.94%), bla CTX M-9 gene 6(17.64%), bla SHV gene: 5(14.70%) and bla TEM gene: 5(14.70%). In summary, analyzed milk samples were found to have a health risk for consumers due to contamination by ESBL producing E. coli, their pathogenicity and treatment failure as a result of antibiotic resistance.
Keywords: Extended Spectrum Beta Lactamase (ESBL), Escherichia coli, Milk, Antibiotic resistance.
Milk is considered as nature’s single most complete food and important part of the diet (Haug et al, 2007). India is the largest milk producing countries in the world with dairy industry playing an important role in the rural economy generating huge self-employment (Batabyal et al., 2018). Contamination of milk can occur due to several causes, moreover; its high nutritive value makes it an ideal medium for the rapid multiplication of bacteria, particularly under unhygienic production and storage at ambient temperatures (Kim et al, 1983; OECD, 2005). Pathogens that have been involved in foodborne outbreaks include Escherichia coli (E. coli), Salmonella and Staphylococcus aureus. The presence of these pathogenic bacteria in milk emerged as major public health concerns, especially for those individuals who still drink raw milk (Riser, 1998). E. coli is mainly responsible for causing diarrhea, urinary tract infection (UTI), hemolytic uremic syndrome (HUS), and hemorrhagic colitis (HC) (Lanjewar et al., 2010).
Extensive reviews of multidrug resistant (MDR) commensal E. coli in animals and its impact on public health has been published recently (Szmolka and Nagy, 2013). One of the most important AMR (Antimicrobial resistance) mechanisms in Enterobacteriaceae family is production of extended-spectrum β-lactamase (ESBL) enzymes. ESBLs are plasmid as well as chromosome mediated and they are the results of the mutation of TEM-1 and TEM-2 and SHV-I genes (Nathisuwan, 2001). ESBL producers are resistant against wide variety of beta
How to cite this article: Paghdar, D., Nayak, J., Mathakiya, R.A., Parmar, B.C., Gida, H.K. and Bhavsar, P.P. (2020). Isolation and molecular characterization of extended spectrum beta lactamase producing Escherichia coli from milk. J. Anim. Res., 10(1): 143-148.
lactam antibiotics including penicilins, 3rd generation cephalosporins and aztreonem. The emergence of ESBL producing E. coli in the food-producing animals and in foods of animal origin is a growing problem worldwide (Geser et al., 2012). Thus, this study was carried out to investigate the microbiological quality and safety of raw cow milk in Anand (Gujarat, India) and around villages.
MATERIALS AND METHODS
Sample collection
A total 150 milk samples were collected from all the milking animals in the dairy cattle farms with at least 10 milking animals in a sterile container in aseptic condition using all the precautions (A- Navali: 25, B- Sai dairy farm: 50, C- Chikhodara: 25 , D- Bedva: 25, E - Mogar: 25). All the collected samples were further sent to the laboratory in an icebox within 1 h of collection and tested immediately upon arrival for further study.
Isolation and Identification
All the samples were first inoculated on MacConkey Agar (MCA) and incubated at 37°C for 24 hours. Plates showing pink colonies (lactose fermenting) were considered as positive and further transferred to Eosin Methylene Blue Agar (EMB) agar. Dark colonies with greenish metallic sheen after 24 hours of incubation at 37°C were considered to be of typical E. coli, were transferred to nutrient agar (NA) slants for further identification and characterization.
Biochemical characterization
Biochemical tests like Indole, Methyl-red, Voges Proskaeur, citrate utilization and TSI tests were employed for identification of E. coli, as per Edwards and Ewing, (1972).
Serotyping of E. coli isolates
Cultures identified as E. coli were serotyped at National Salmonella and Escherchia Centre (NSEC), Central Research Institute (CRI), Kasauli (Himachal Pradesh, India).
Antibiotic sensitivity test
Phenotypic confirmation of ESBL producing E. coliisolates was done using in vitro antibiotic sensitivity test by disc diffusion method using the discs supplied by HiMedia Laboratories Pvt. Ltd., Mumbai (India). Isolates were subjected to antimicrobial sensitivity tests against 6 antibiotics of 3rd generation cephalosporins viz.; Cefoxitin (30 μg), Ceftriaxone (30 μg), Ceftazidime (30 μg), Cefpodoxim (30 μg), Aztrionem (30 μg) and Ceftazidime + Clavulanic acid (30/10 μg). A difference of > 5 mm between the zone diameters of ceftazidme and Ceftazidime + Clavulanic acid disc is measured to phenotypically confirm the ESBL production by the E. coli isolates under study.
Epsilometer test
All the E. coli isolates were tested again for phenotypic confirmation of ESBL producing E. coli by E test using ESBL strip which carries two gradients; on the one end, ceftazidime and on the opposite end ceftazidime plus clavulanic acid. MIC is interpreted as the point of intersection of the inhibition ellipse with the E test strip edge. Ratio of ceftazidime MIC and ceftazidime clavulanic acid MIC ≥ 8 indicates the presence of ESBL (Bush and Jacoby, 1995).
Table 1: Primer sequences for blaCTX M-3’blaCTX M-9’blaSHV’ blaTEM
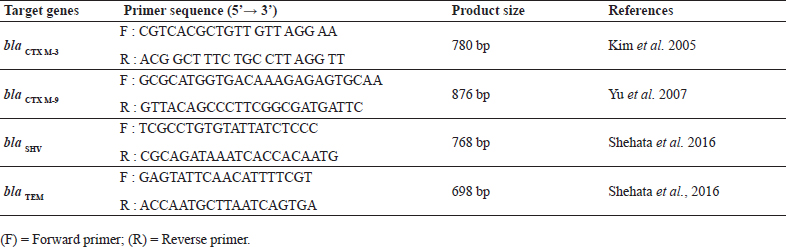
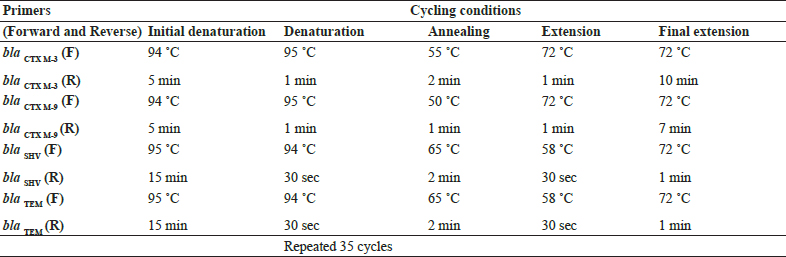
Table 2: Steps and conditions of thermal cycling for different primer pairs in PCR
Polymerase chain reaction
Genotypic confirmation of ESBL genes was done for all the phenotypically confirmed ESBL producing E. coliusing the PCR protocols separately standardized for the detection of different ESB genes viz.; blaCTXM-3 , blaCTX M-9, blaSHV, blaTEM. The PCR was standardized for the detection of four genes following the methodology as described in table 2 with suitable modifications (Kim et al., 2005, Yu et al., 2007 and Shehata et al., 2016).
RESULTS AND DISCUSSION
Isolation and Identification
A total 94 (62.66%) E. coli isolates were recovered from 150 raw milk samples which were collected from Anand and around villages (Navali-17, Sai dairy farm-32, Chikhodara-14, Bedva-16, Mogar-15). All the positive isolates showed typical characteristics of E. coli viz., greenish metallic sheen on EMB agar (Fig. 2), positive indole and methyl red, negative VP and citrate utilization test and yellow slant, yellow butt and no H2S production on TSI agar.
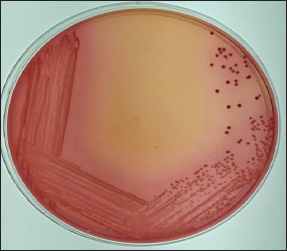
Fig. 1 : Lactose fermenting pink colonies of gram negative bacteria on MacConkey agar plate
Fig. 2 : Greenish metallic sheen producing colonies of E. coli on Eosin Methylene Blue agar plate
Serotyping of E. coli isolates
Cultures identified as E. coli were serotyped at National Salmonella and Escherchia Centre (NSEC), Central Research Institute (CRI), Kasauli (Himachal Pradesh, India). Out of 94 E. coli isolates 60 (63%), 28 (29%) isolates were unypable (UT) and 6 (6%) were found to be rough. A total 14 different serotypes were found in the present study namely; O83, O49, O157, O103, O22, O34, O26, O145, O146, O2, O141, O119, O84 and O120.
Antibiotic sensitivity test
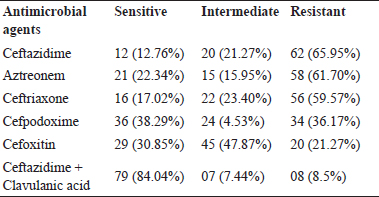
In vitro antimicrobial susceptibility test (ABST) was performed for all 94 E. coli isolates by disk diffusion method and results were as shown in Table 3. Maximum isolates (84.04%) were sensitive to antibiotic Ceftazidime + Clavulanic acid combination while maximum isolates (65.95%) were resistance to Ceftazidime. A total 34 (36%) isolates out of 94 isolated were found positive as ESBL producing E. coli when the difference in the inhibition zone was measured between ceftazidime and ceftazidime + cavulanic acid; organisms giving the difference in inhibition zone ≥ 5 were considered as positive ESBL producers.
Table 3: In vitro antimicrobial drug resistance pattern of E. coliisolates
Epsilometer test
Out of 94 E. coli isolates, 34 (36.00%) isolates were phenotypically confirmed as ESBL producer through E test. Here for the interpretation ESBL producing E. coligive ratio of MIC of Mix +/ MIC of Mix ≥ 8 is considered as positive ESBL producers.
Polymerase chain reaction
Out of 34 phenotypically confirmed ESBL producing E. coli isolates, 18 (52.94%) isolates were positive for bla CTX M- 3 gene, 6 (17.64%) isolates were positive for bla CTX M-9, 5 (l4.70%) isolates were positive bla SHV & also 5 (14.70%) isolates were positive bla TEM genes.
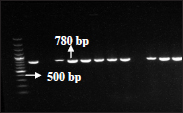
Fig. 3: PCR amplified product (780bp) for blaCTX M-3 gene

Fig. 4: PCR amplified product (876bp) for blaCTX M-9 gene
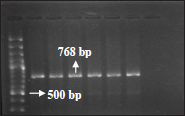
Fig. 5: PCR amplified product (768bp) for bla SHV gene
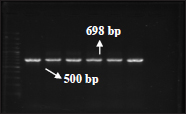
Fig. 6: PCR amplified product (698bp) for bla TEM gene
Here, in the present study 62.66% isolates from raw milk samples revealed characteristic features of E. coli which is in agreement with the prior findings; 65% by Chye et al.(2004), 52% by Virpari et al. (2013), 65% by Soomro et al. (2002), 63% by Uddin et al. (2011).
The serotypes reported by the National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, were also supported by Manna et al. (2006), Miszczycha et al. (2013), Hardik et al. (2017) who reported E. coliserotype O157, O83, O84, O145 and O103 in their studies. Elhadidy & Mohammed (2013) and Blanco et al. (2004) reported the presence of serotypes O22, O120, O2, O141 O119, O34 and O49 from E. coli isolated from raw milk samples which is in agreement to the present study.
Gashe et al. (2018) reported 60% resistance against ceftazidime and 59% against ceftriaxone in a prior study. Xu et al. (2018) reported 20% resistance against cefoxitin, Jena et al. (2018) reported 61% resistance against aztreonem and 36% against cefpodoxime in theie studies. Bhattacharya et al. (2015) observed higher degree (84.04%) of sensitivity of E. coli isolates when Ceftazidime + Clavulanic acid combination was used. Results of all these studies support the findings of the present study.
In the present study, total 34 (36%) E. coli isolates were found positive through E test which is in agreement to prior study by Cormican et al. (1996) where 82 (36%) E. coli isolates were found positive as ESBL producers out of 225.
In the present study, 18 (52%), 6 (17%), 5 (14%) and 5 (14%) isolates were positive for bla CTX M-3, bla CTX M-9, bla TEM and bla SHV genes respectively. Similar results were reported by Tekiner and Ozpinar (2016); where 53% prevalence of bla CTX M-3, 12% for bla TEM & bla SHV genes, and 16% for bla CTX M-9 was observed.
CONCLUSION
Analysed raw milk samples poses high health risk to the consumers because of such high prevalence (94%) of E. coli from milk. A high degree of the resistance (80%) was observed by ESBL producing E. coli towards beta lactam antibiotics which can be considered a reason of the treatment failure and economic losses thus it rises great public health concerns. It is suggested to administer beta lactam antibiotic along with sucidal inhibitors like Clavulanic acid, Tazobactam, and Salbactam for cost effective treatment and to meet the purpose of public health.
Acknowledgements
Authors are thankful to department of Veterinary Pubic Health & Epidemiology, Anand Veterinary College, A.A.U., Anand.
REFERENCES
Batabyal, K., Banerjee, A., Pal, S., Dey, S., Joardar, S.N., Samanta, I., Isore, D.P. and Singh, A.D. 2018. Detection, characterization, and antibiogram of extended-spectrum beta-lactamase Escherichia coli isolated from bovine milk samples in West Bengal, India, Vet. World., 11: 1423-1427.
Bhattacharya, C., Das, N. and Pal, D. 2015. Detection of extended spectrum Betaactamase (ESBL) producing bacteria from meat and meat products in Kolkata, India. J. Dental. Medical. Sci., 14: 52-55.
Blanco, M., Blanco, J. E., Mora, A., Dahbi, G., Alonso, M. P., González, E. A. and Blanco, J. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)- producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-ε). J. Clin. Microbiol., 42: 645-651.
Bush, K., Jacoby, G.A. and Medeiros, A.A. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents. Chemother.,39: 1211-1233.
Chye, F.Y., Abdullah, A. and Ayob, M K. 2004. Bacteriological quality and safety of raw milk in Malaysia. Food. Microbiol., 21: 535-541.
Cormican, M.G., Marshall, S.A. and Jones, R.N. 1996. Detection of extended-spectrum beta-lactamase (ESBL)-producing strains by the Etest ESBL screen. J. Clin. Microbiol., 34: 1880-1884.
Edwards, PR. and Ewing, W.H. 1972. The genus Salmonella. Identifica. Enterobacter., 146-207.
Elhadidy, M. and Mohammed, M.A. 2013. Shiga toxin- producing Escherichia coli from raw milk cheese in Egypt: prevalence, molecular characterization and survival to stress conditions. Lett. Appl. Microbiol., 56: 120-127.
Gashe, F., Mulisa, E., Mekonnen, M. and Zeleke, G. 2018. Antimicrobial resistance profile of different clinical isolates against third-generation cephalosporins. Int. J. Pharm., 2018.
Geser, N., Stephan, R. and Hächler, H. 2012. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Research, 8: 21.
Hardik, P., Kalyani, I.H., Bariya, A., Dodiya, V, Sakhare, P. and Sharma, K. 2017. Isolation, characterization, serotyping and antibiogram studies of E. coli collected from diarrhoeic neonatal kids. Int. J. Microbiol., 9: 854-856.
Haug, A., H0stmark, A.T. and Harstad, O.M. 2007. Bovine milk in human nutrition-a review. Lipids Health Dis., 6: 25.
Jena, J., Debataa, N.K., Sahoob, R.K., Gaurb, K. and Subudhib, M. 2018. Molecular characterization of extended spectrum β-lactamase-producing Enterobacteriaceae strains isolated from a tertiary care hospital. Microbial Pathog., 115: 112116.
Kim, H. 1983. Off-tastes in raw and reconstituted milk. Food ȁ Agriculture Org. (Vol. 35).
Kim, J., Lim, Y.M., Rheem, I., Lee, Y., Lee, J.C., Seol, S.Y. and Cho, D.T. 2005. CTX-M and SHV-12 β-lactamases are the most common extended-spectrum enzymes in clinical isolates of Escherichia coli and Klebsiella pneumoniae collected from 3 university hospitals within Korea. FEMS Microbiol. Lett., 245: 93-98.
Lanjewar, M., De Anuradha, S. and Mathur, M. 2010. Diarrheagenic E. coli in hospitalized patients: special reference to Shiga-like toxin producing Escherichia coli. Indian J. Pathol. Microbiol., 53: 75.
Manna, S.K., Brahmane, M.P., Manna, C., Batabyal, K. and Das, R. 2006. Occurrence, virulence characteristics and antimicrobial resistance of Escherichia coli O157 in slaughtered cattle and diarrhoeic calves in West Bengal, India. Lett. Appl. Microbiol., 43: 405-409.
Miszczycha, S.D., Perrin, F., Ganet, S., Jamet, E., Tenenhaus- Aziza, F., Montel, M. C. and Thevenot-Sergentet, D. 2013. Behavior of different Shiga toxin-producing Escherichia coli serotypes in various experimentally contaminated raw-milk cheeses. Appl. Environ. Microbiol., 79: 150-158.
Nathisuwan, S., Burgess D.S. and Lewis, J.S. 2001. ESBLs: Epidemiology, Detection and Treatment. Pharmacotherapy, 21: 920-928.
OECD. 2005. Dairy policy reform and trade liberalization. Organization for Economic Cooperation and Development (OECD publishing), 98.
Riser, E.T. 1998. Public health concerns. In: edited by Marth EH and Steele JL. Appl. Dairy Microbiol., 263-403.
Shehata, M.E., EL - Sherbiny, G. M., Mohamed, A.H. and Shafik, H.M. 2016. Molecular and Phenotypic Characterization of some antimicrobial resistance genes in Escherichia coli isolated from human and broiler chickens. Int. J. Curr.t Microbiol. Appl. Sci., 5: 953-965.
Soomro, A.H., Arain, M.A., Khaskheli, M. and Bhutto, B. 2002. Isolation of Escherichia coli from raw milk and milk products in relation to public health sold under market conditions at Tandojam. Pak. J. Nutr., 1: 151-152.
Szmolka, A. and Nagy, B. 2013. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol., 4: 258.
Tekiner, I.H. and Ozpinar, H. 2016. Occurance amd characteristics of extended spectrum beta-lactamase- producing Enterobacteriaceae from foods of animal origin. Braz. J. Microbiol., 47: 444-451.
Uddin, M.A., Motazzim-ul-Haque, H.M. and Noor, R. 2011. Isolation and identification of pathogenic Escherichia coli, Klebsiella spp. and Staphylococcus spp. in raw milk samples collected from different areas of Dhaka City, Bangladesh. S. J. Microbiol., 1: 19-23.
Virpari, PK., Nayak, J.B., Thaker, H.C. and Brahmbhatt, M.N. 2013. Isolation of pathogenic Escherichia coli from stool samples of diarrhoeal patients with history of raw milk consumption. Vet. World, 6: 659-663.
Xu, Y., Sun, H., Bai, X., Fu, S., Fan, R. and Xiong, Y. 2018. Occurrence of multidrug-resistant and ESBL-producing atypical enteropathogenic Escherichia coli in China. Gut Pathog., 10: 8.
Yu, Yunsong, Shujuan J., Yagang C., Weilin Z., Zeqing W., Lanjuan L. and Yilin M. 2007. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J. Infect., 54: 53-57.